DEVICE='cpu'
E=49
model_name = 'sbm' + f'_E{E}'Loading checkpoint: ./models/sbm_E49.tar
on device: cpuThe goal of this tutorial is to generate new trajectories from the trained SPIVAE and examine their attributes to evaluate the model’s strengths and drawbacks.
We load the model to characterize from its checkpoint.
Loading checkpoint: ./models/sbm_E49.tar
on device: cpuWe can check the parameters of the data and the model.
{'path': '../../data/raw/', 'model': 'sbm', 'N': 476, 'T': 400, 'D': array([1.00000000e-05, 2.15443469e-05, 4.64158883e-05, 1.00000000e-04,
2.15443469e-04, 4.64158883e-04, 1.00000000e-03, 2.15443469e-03,
4.64158883e-03, 1.00000000e-02]), 'alpha': array([0.2 , 0.28, 0.36, 0.44, 0.52, 0.6 , 0.68, 0.76, 0.84, 0.92, 1. ,
1.08, 1.16, 1.24, 1.32, 1.4 , 1.48, 1.56, 1.64, 1.72, 1.8 ]), 'N_save': 1000, 'T_save': 400, 'seed': 0, 'valid_pct': 0.2, 'bs': 256} {'o_dim': 399, 'nc_in': 1, 'nc_out': 6, 'nf': [16, 16, 16, 16], 'avg_size': 16, 'encoder': [200, 100], 'z_dim': 6, 'decoder': [100, 200], 'beta': 0.005, 'in_channels': 1, 'res_channels': 16, 'skip_channels': 16, 'c_channels': 6, 'g_channels': 0, 'res_kernel_size': 3, 'layer_size': 4, 'stack_size': 1, 'out_distribution': 'Normal', 'num_mixtures': 1, 'use_pad': False, 'model_name': 'SPIVAE'}As the latent neurons’ interpretation is complex, we feed the model with known trajectories and use their latent representation for the generation. We load the test set generated in the data docs or the analysis tutorial.
(True, False, 256, 'cpu')We can see the mean loss on a batch.
RF: 32 bs: 256
Current mean loss on a batch: -2.2795233726501465We generate new displacements from latent representation of the inputs.
ds_in,preds,ds_targs = learn.get_preds(with_input=True,) # predicts in validation dataset
alphas_items = learn.dls.valid.items[:,0]
Ds_items = learn.dls.valid.items[:,1]
ds_in_labels = learn.dls.valid.items[:,:2]
u_a=np.unique(alphas_items,)
u_D=np.unique(Ds_items,)
alphas_idx = [np.flatnonzero(alphas_items==a) for a in u_a]
Ds_idx = [np.flatnonzero(Ds_items==D) for D in u_D]
intersect_idx = np.array([[reduce(partial(np.intersect1d,assume_unique=True),
(alphas_idx[i],Ds_idx[j]))
for j,D in enumerate(u_D)]
for i,a in enumerate(u_a)], dtype=object)
alphas_idx_flat = [item for sublist in alphas_idx for item in sublist]
Ds_idx_flat = [item for sublist in Ds_idx for item in sublist]
pred, mu, logvar, c = preds
print(ds_in.shape, learn.dls.valid.items.shape)
print(L(map(len, alphas_idx)), len(alphas_idx_flat), L(map(len, Ds_idx)), len(Ds_idx_flat))torch.Size([67200, 1, 399]) (67200, 401)
[3181, 3202, 3224, 3174, 3202, 3183, 3203, 3203, 3196, 3215, 3184, 3196, 3213, 3190, 3207, 3210, 3171, 3242, 3188, 3212, 3204] 67200 [16844, 16738, 16802, 16816] 67200We choose the number of samples for each parameter combination, the number of samples for each processed input, and the batch size for generation.
We sample from the model and save it for each parameter combination.
for a_idx,a in enumerate(ds_args["alpha"]):
for D_idx,D in enumerate(ds_args["D"]):
fname = "../../data/gen/"+model_name+f'_disp_gen_a{a:.3g}D{D:.3g}'.replace('.','')+'.npz'
if not os.path.exists(fname):
os.makedirs('../../data/gen/', exist_ok=True)
indices = (a_idx,D_idx)
disp_gen, logits = learn.model.sample_batch(total_samples, bs=bs,
c=c[intersect_idx[indices][:trajs_per_aD]].repeat(samples_per_c,1,1))
disp_gen = disp_gen.squeeze().detach().cpu().numpy()
np.savez_compressed(fname, disp_gen=disp_gen)
print("Saved at:", fname)
print("with dimensions", disp_gen.shape)Saved at: ../../data/gen/sbm_E49_disp_gen_a02D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a02D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a02D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a02D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a028D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a028D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a028D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a028D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a036D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a036D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a036D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a036D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a044D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a044D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a044D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a044D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a052D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a052D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a052D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a052D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a06D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a06D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a06D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a06D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a068D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a068D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a068D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a068D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a076D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a076D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a076D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a076D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a084D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a084D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a084D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a084D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a092D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a092D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a092D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a092D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a1D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a1D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a1D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a1D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a108D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a108D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a108D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a108D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a116D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a116D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a116D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a116D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a124D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a124D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a124D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a124D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a132D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a132D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a132D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a132D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a14D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a14D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a14D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a14D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a148D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a148D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a148D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a148D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a156D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a156D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a156D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a156D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a164D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a164D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a164D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a164D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a172D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a172D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a172D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a172D001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a18D1e-05.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a18D00001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a18D0001.npz
with dimensions (6000, 200)Saved at: ../../data/gen/sbm_E49_disp_gen_a18D001.npz
with dimensions (6000, 200)We analyze the diffusion properties of the displacements by means of standard statistical techniques. The main approach consists in characterizing ensembles of displacements with the same properties, averaging in time, samples, or both.
We first load the generated displacements for the combination of parameters we want.
As a fast check of the generation, we can see if the displacements mean and variance are the expected. For each parameter combination, we observe the mean of generated displacements is inside the limits we saw with the input.
For the variance, we observe a correct reproduction of the scaling with a small deviation, as noted in the analysis.
for j,D in enumerate(ds_args["D"]):
plt.figure()
for i,a in enumerate(ds_args["alpha"][::5]):
k = f'{a:.3g}'+f',{D:.3g}'
# original time
t_full = np.arange(1,disp_gen[k].shape[-1]+1)
# shifted
t_shift = model.receptive_field+1
t_gen = t_full + t_shift
# scaling to compensate the time generated by the machine starting at RF+1
# time scale factors: ((1+t_full)/(t_shift+t_full))**(a-1.)
t_scaling = ((1+t_full)/(t_gen))**(a-1.)
x = sig2D(disp_gen[k].squeeze().std(0))
x = x*t_scaling
plt.semilogy(x, '-', c=f'C{i}', label=f'{a:.3g}');
plt.plot(a*D*np.arange(1,x.shape[-1])**(a-1.), '--')
plt.axhline(D, c='k', lw=1, ls='--', alpha=0.7);
plt.xlabel(r'$t$');plt.ylabel(r'$D_{gen}$');
h,l = plt.gca().get_legend_handles_labels()
plt.legend(h[::-1],l[::-1],title=r'$\alpha$', bbox_to_anchor=(1,1));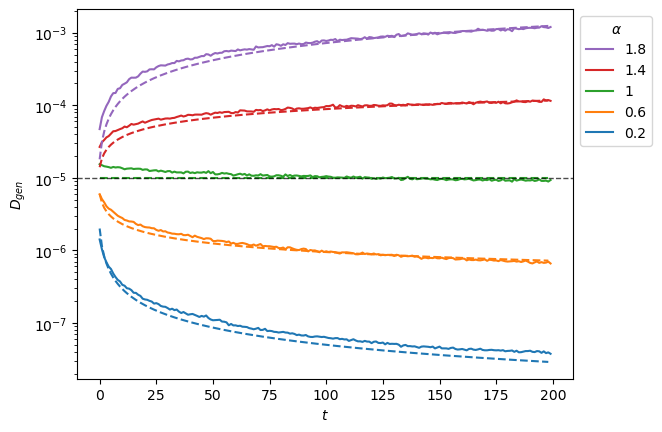
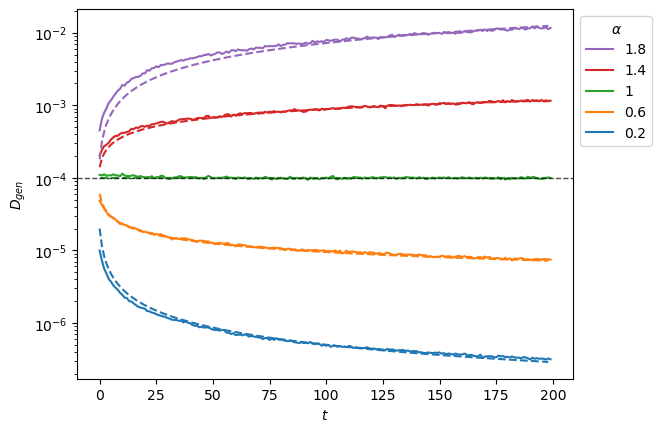
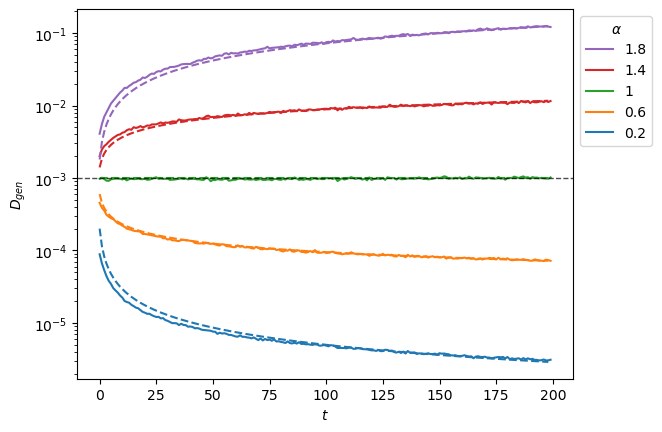
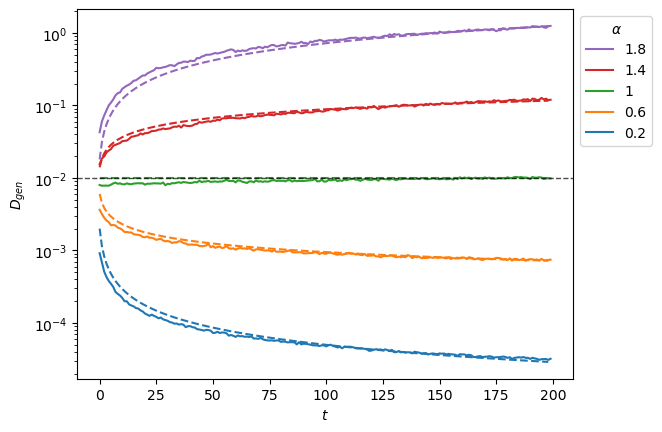
To extract the anomalous exponent \(\alpha\) from SBM trajectories, one needs the time ensemble average mean squared displacement (TEA-MSD). As SBM is weakly non-ergodic, the TA-MSD along different time lags will not work as it did for FBM. We instead make the TA-MSD for one time lag and take the ensemble average.
def TAMSD_t_lag(traj, t_lag):
t_t_lag = len(traj) - t_lag
tamsd = np.zeros(t_t_lag, dtype=np.float64)
for t_ in range(t_t_lag): tamsd[t_] = (traj[t_] - traj[t_ + t_lag])**2
tamsd /= (t_t_lag)
return tamsd
def TEAMSD_t_lag(trajs, t_lag):
ttt = np.zeros((trajs.shape[0], trajs.shape[-1]-t_lag), dtype=np.float64)
for i in range(trajs.shape[0]): # prange
ttt[i] = TAMSD_t_lag(trajs[i], t_lag)
return tttdef get_alpha_from_sbm_trajs(trajs, # (N,T)
t_lag=2, perc=.55):
teamsd = TEAMSD_t_lag(trajs,t_lag).mean(0)
log_teamsd = np.log(teamsd)
# if two timesteps have the same value, tamsd_=0 and log is -inf and polyfit is nan
log_teamsd[~np.isfinite(log_teamsd)]=-27
log_t = np.log(np.arange(1,teamsd.shape[-1]+1))
last_perc = int(log_teamsd.shape[-1]*perc)
return 1+np.polyfit(log_t[-last_perc:], log_teamsd[-last_perc:], 1)[0]cmap_D = matplotlib.colors.ListedColormap(matplotlib.colormaps['Greens'](np.linspace(0.3,1,150)))
norm_D = matplotlib.colors.LogNorm(vmin=3.16e-6, vmax=3.16e-2)
sm_D_4 = plt.cm.ScalarMappable(cmap=cmap_D, norm=norm_D)
for j,D in enumerate(ds_args["D"]):
x = ds_args["alpha"]
y = exponents[:,j]
plt.plot([0,1,2], 'gray', alpha=0.7, zorder=-1);
plt.plot(x,y, c=cmap_D(norm_D(D)),label=f'{D:.3g}');
ax = plt.gca(); ax.grid();
ax.set_xlabel(r'$\alpha$', ha='center', va='top',);
ax.set_ylabel(r'$\alpha_{g}$', rotation=0, ha='right', va='center');
ax.set_xlim(0,2); ax.set_ylim(0,2);
ax.locator_params(nbins=3);
ax.set_xticks([0.2,1,1.8],[0.2,'1',1.8])
cb = plt.colorbar(sm_D_4, ax=ax,pad=0, aspect=9,shrink=0.3, anchor=(-2,0.125));
cb.ax.set_ylabel(r'$D_0$',ha='left', va='center', rotation=0, )
cb.ax.set_yscale('log')
cb.ax.set_yticks([1e-5,1e-2], [r'$10^{-5}$',r'$10^{-2}$']);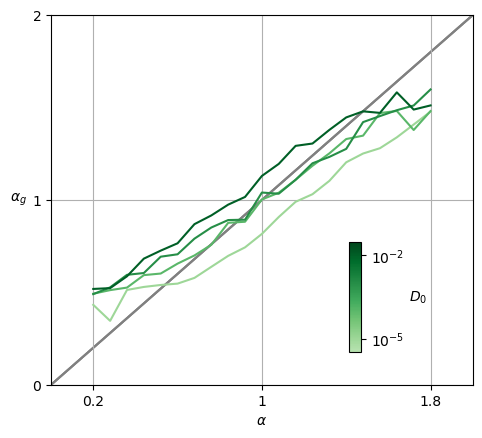
The estimated exponent \(\alpha_g\) is very close to the ground truth of the input for different values of \(D_0\).