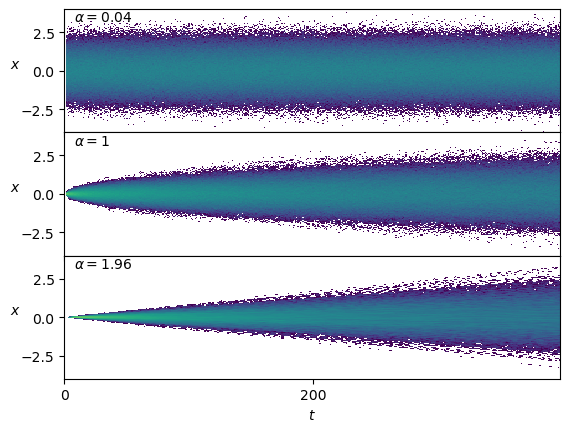
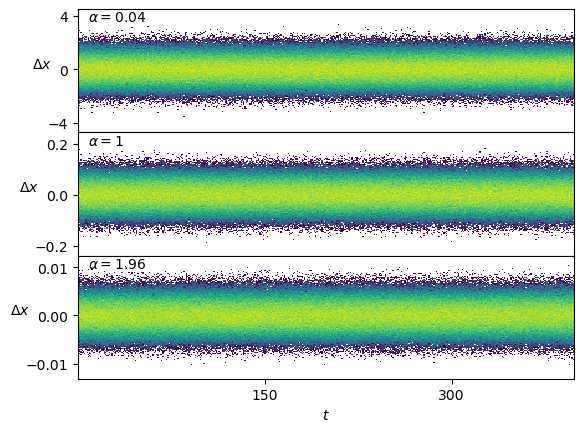
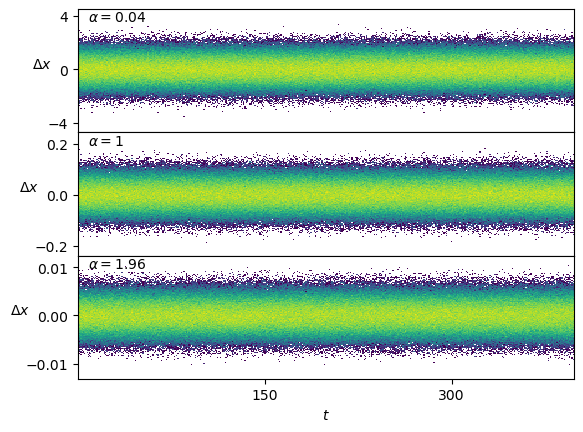
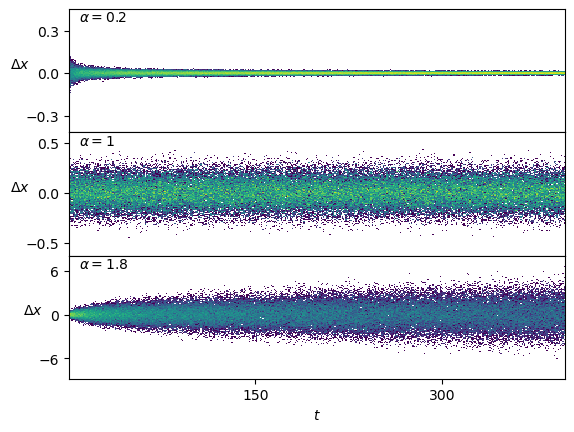
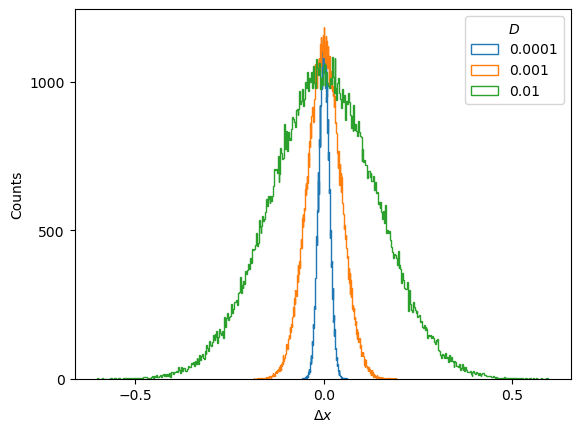
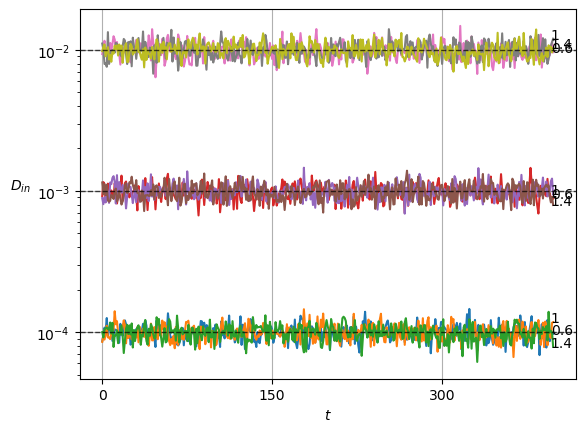
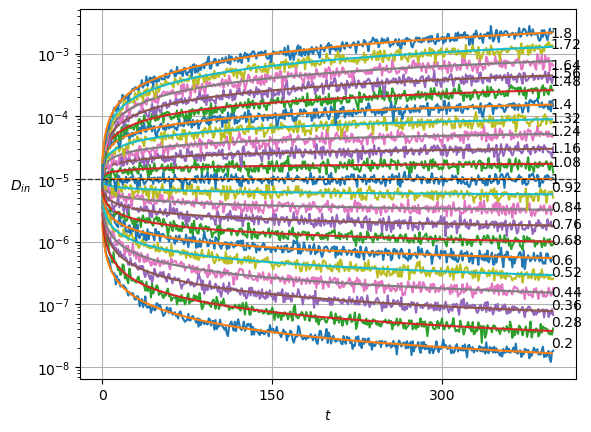
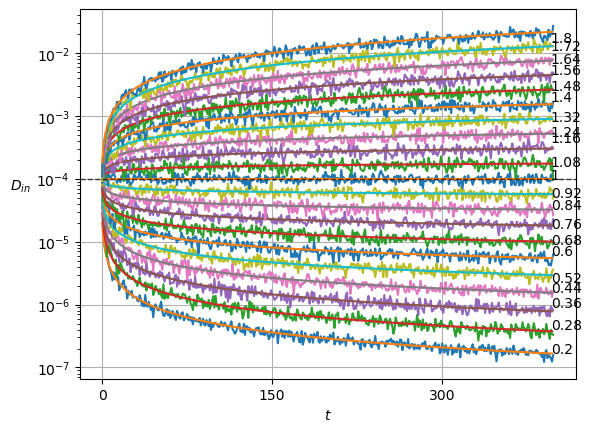
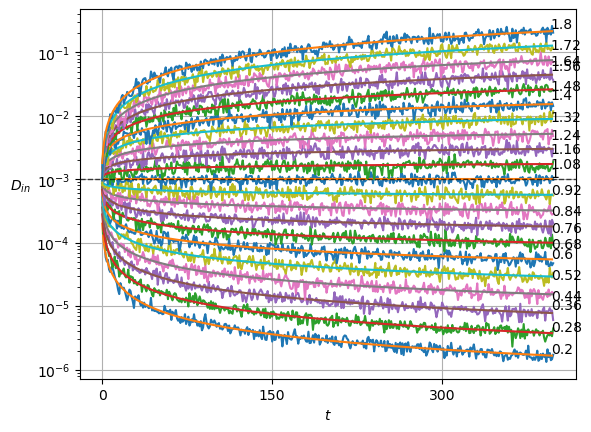
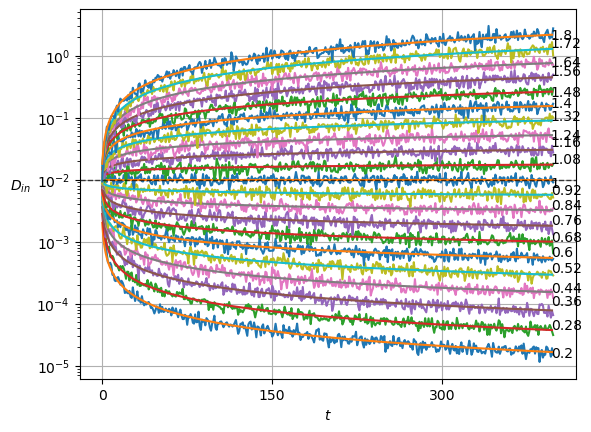
alphas_train = np.linspace(0.2,1.8,21)
alphas_test = np.linspace(0.04,1.96,49)
print(f'{alphas_train=}')
print(f'{alphas_test=}')alphas_train=array([0.2 , 0.28, 0.36, 0.44, 0.52, 0.6 , 0.68, 0.76, 0.84, 0.92, 1. ,
1.08, 1.16, 1.24, 1.32, 1.4 , 1.48, 1.56, 1.64, 1.72, 1.8 ])
alphas_test=array([0.04, 0.08, 0.12, 0.16, 0.2 , 0.24, 0.28, 0.32, 0.36, 0.4 , 0.44,
0.48, 0.52, 0.56, 0.6 , 0.64, 0.68, 0.72, 0.76, 0.8 , 0.84, 0.88,
0.92, 0.96, 1. , 1.04, 1.08, 1.12, 1.16, 1.2 , 1.24, 1.28, 1.32,
1.36, 1.4 , 1.44, 1.48, 1.52, 1.56, 1.6 , 1.64, 1.68, 1.72, 1.76,
1.8 , 1.84, 1.88, 1.92, 1.96])