model_args = dict(# VAE #########################
o_dim=400,
nc_in=1, nc_out=6,
nf=[16]*4,
avg_size=16,
encoder=[200,100],
z_dim=6,
decoder=[100,200],
beta=0,
# WaveNet ########
in_channels=1,
res_channels=16,skip_channels=16,
c_channels=6,
g_channels=0,
res_kernel_size=3,
layer_size=4, # 6
stack_size=1,
out_distribution= "Normal",
num_mixtures=1,
use_pad=False,
model_name = 'SPIVAE',
)
model = VAEWaveNet(**model_args)Model
The architecture of the interpretable autoregressive \(\beta\)-VAE works in the following manner: Given the displacements \(\mathbf{\Delta x}(t)\) of a diffusion trajectory, the encoder (orange) compresses them into an interpretable latent space (blue), in which few neurons (dark blue) represent physical features of the input data while others are noised out (light blue). An autoregressive decoder (green) generates from this latent representation the displacements \(\mathbf{\Delta x}'(t)\) of a new trajectory recursively, considering a certain receptive field RF (light green cone). 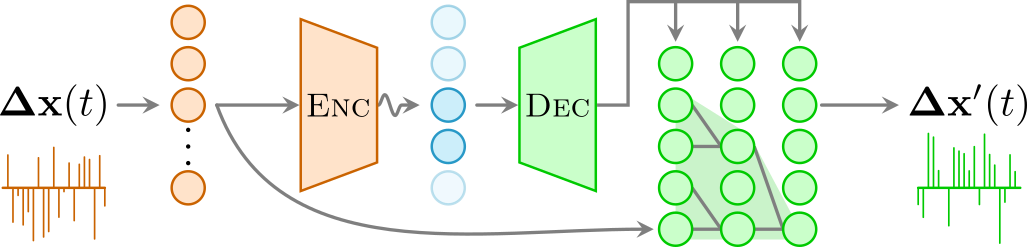
Initialization
As the architecture can be quite deep, a careful initialization is needed (see weight_init function in the model class). We initialize the weights with normal Kaiming init in fan_out mode, taking into account that we use the nonlinear activation function ReLU.
init_cnn
def init_cnn(
m
):
Initialize weights with kaiming normal in fan_out mode and bias to 0
VAE
We implement a 1D convolutional variational autoencoder.
The latent neurons are probabilistic, i.e., they are sampled following a distribution. The reparameterization trick provides the means to allow backpropagation by externalizing the sampling noise.
reparameterize
def reparameterize(
mu, # Mean of the normal distribution. Shape (`batch_size`, `latent_dim`)
logvar, # Diagonal log variance of the normal distribution. Shape (`batch_size`, `latent_dim`)
): # Sampled latent `z` tensor as $z=\epsilon\sigma+\mu$
Samples from a normal distribution using the reparameterization trick.
We also take into account the sizes after n convolutions are applied to automate the model construction.
output_size_after_n_convt
def output_size_after_n_convt(
n, input_size, kernel_size, stride:int=1, padding:int=0, output_padding:int=0, dilation:int=1
):
output_size_convt
def output_size_convt(
input_size, kernel_size, stride:int=1, padding:int=0, output_padding:int=0, dilation:int=1
):
output_size_after_n_conv
def output_size_after_n_conv(
n, input_size, kernel_size, stride:int=1, padding:int=0, dilation:int=1
):
output_size_conv
def output_size_conv(
input_size, kernel_size, stride:int=1, padding:int=0, dilation:int=1
):
View
def View(
size
):
Use as (un)flattening layer
VAEConv1d
def VAEConv1d(
nf, # number of filters
encoder, # list of Encoder's dense layers sizes
decoder, # list of Decoder's dense layers sizes
o_dim:int, # input size (T)
nc_in:int=1, # number of input channels
nc_out:int=6, # number of output channels
z_dim:int=6, # number of latent neurons
beta:int=0, # weight of the KLD loss
avg_size:int=24, # output size of the pooling layers
kwargs:VAR_KEYWORD
):
1-dimensional convolutional VAE architecture
VAE + WaveNet
We implement an extensible version of a VAE with WaveNet as the autoregressive decoder.
sample_from_mix_gaussian
def sample_from_mix_gaussian(
y, # Mixture of Gaussians parameters. Shape (B x C x T)
log_scale_min:float=-12.0, # Log scale minimum value.
In many other implementations this variable is never used.
):
Sample from (discretized) mixture of gaussian distributions
DilatedCausalConv1d
def DilatedCausalConv1d(
mask_type, in_channels, out_channels, kernel_size:int=2, dilation:int=1, bias:bool=True, use_pad:bool=True
):
Dilated causal convolution for WaveNet
ResidualBlock
def ResidualBlock(
res_channels, skip_channels, kernel_size, dilation, c_channels:int=0, g_channels:int=0, bias:bool=True,
use_pad:bool=True
):
Residual block with conditions and gate mechanism
VAEWaveNet
def VAEWaveNet(
in_channels:int=1, # input channels
res_channels:int=16, # residual channels
skip_channels:int=16, # skip connections channels
c_channels:int=6, # local conditioning
g_channels:int=0, # global conditioning
out_channels:int=1, # output channels
res_kernel_size:int=3, # kernel_size of residual blocks dilated layers
layer_size:int=4, # Largest dilation is 2^layer_size
stack_size:int=1, # number of layers stacks
out_distribution:str='normal', discrete_channels:int=256, num_mixtures:int=1, use_pad:bool=False,
weight_norm:bool=False, kwargs:VAR_KEYWORD
):
VAE with autoregressive decoder
We can create a model by specifying its parameters in a dict.
Printing the model object will reveal the declared layers.
modelVAEWaveNet(
(vae): VAEConv1d(
(encoder): Sequential(
(0): Conv1d(1, 16, kernel_size=(3,), stride=(1,))
(1): ReLU(inplace=True)
(2): Conv1d(16, 16, kernel_size=(3,), stride=(1,))
(3): ReLU(inplace=True)
(4): Conv1d(16, 16, kernel_size=(3,), stride=(1,))
(5): ReLU(inplace=True)
(6): Conv1d(16, 16, kernel_size=(3,), stride=(1,))
(7): ReLU(inplace=True)
(8): AdaptiveConcatPool1d(
(ap): AdaptiveAvgPool1d(output_size=16)
(mp): AdaptiveMaxPool1d(output_size=16)
)
(9): View()
(10): Linear(in_features=512, out_features=200, bias=True)
(11): ReLU(inplace=True)
(12): Linear(in_features=200, out_features=100, bias=True)
(13): ReLU(inplace=True)
(14): Linear(in_features=100, out_features=12, bias=True)
)
(decoder): Sequential(
(0): Linear(in_features=6, out_features=100, bias=True)
(1): ReLU(inplace=True)
(2): Linear(in_features=100, out_features=200, bias=True)
(3): ReLU(inplace=True)
(4): Linear(in_features=200, out_features=512, bias=True)
(5): ReLU(inplace=True)
(6): View()
)
(convt): Sequential(
(0): ConvTranspose1d(16, 16, kernel_size=(3,), stride=(1,))
(1): ReLU(inplace=True)
(2): ConvTranspose1d(16, 16, kernel_size=(3,), stride=(1,))
(3): ReLU(inplace=True)
(4): ConvTranspose1d(16, 16, kernel_size=(3,), stride=(1,))
(5): ReLU(inplace=True)
(6): ConvTranspose1d(16, 6, kernel_size=(3,), stride=(1,))
(7): ReLU(inplace=True)
)
)
(init_conv): Conv1d(1, 16, kernel_size=(1,), stride=(1,))
(causal): DilatedCausalConv1d(
(conv): Conv1d(16, 16, kernel_size=(2,), stride=(1,))
)
(res_stack): ModuleList(
(0): ResidualBlock(
(dilated): DilatedCausalConv1d(
(conv): Conv1d(16, 32, kernel_size=(3,), stride=(1,))
)
(conv_c): Conv1d(6, 32, kernel_size=(1,), stride=(1,), bias=False)
(conv_res): Conv1d(16, 16, kernel_size=(1,), stride=(1,))
(conv_skip): Conv1d(16, 16, kernel_size=(1,), stride=(1,))
)
(1): ResidualBlock(
(dilated): DilatedCausalConv1d(
(conv): Conv1d(16, 32, kernel_size=(3,), stride=(1,), dilation=(2,))
)
(conv_c): Conv1d(6, 32, kernel_size=(1,), stride=(1,), bias=False)
(conv_res): Conv1d(16, 16, kernel_size=(1,), stride=(1,))
(conv_skip): Conv1d(16, 16, kernel_size=(1,), stride=(1,))
)
(2): ResidualBlock(
(dilated): DilatedCausalConv1d(
(conv): Conv1d(16, 32, kernel_size=(3,), stride=(1,), dilation=(4,))
)
(conv_c): Conv1d(6, 32, kernel_size=(1,), stride=(1,), bias=False)
(conv_res): Conv1d(16, 16, kernel_size=(1,), stride=(1,))
(conv_skip): Conv1d(16, 16, kernel_size=(1,), stride=(1,))
)
(3): ResidualBlock(
(dilated): DilatedCausalConv1d(
(conv): Conv1d(16, 32, kernel_size=(3,), stride=(1,), dilation=(8,))
)
(conv_c): Conv1d(6, 32, kernel_size=(1,), stride=(1,), bias=False)
(conv_res): Conv1d(16, 16, kernel_size=(1,), stride=(1,))
(conv_skip): Conv1d(16, 16, kernel_size=(1,), stride=(1,))
)
)
(out_conv): Sequential(
(0): ReLU(inplace=True)
(1): Conv1d(16, 16, kernel_size=(1,), stride=(1,))
(2): ReLU(inplace=True)
(3): Conv1d(16, 9, kernel_size=(1,), stride=(1,))
)
)Training example
With the data and the model, we can already start training.
DEVICE= 'cpu' # 'cuda'
print(DEVICE)cpuN=6_000Ds = np.linspace(2e-5,2e-2,5)
alphas = np.linspace(0.2,1.8,9)
n_alphas,n_Ds = len(alphas), len(Ds)
ds_args = dict(path="../../data/raw/", model='fbm', # 'sbm'
N=int(N/n_alphas/n_Ds*2), T=400,
D=Ds, alpha=alphas,seed=0,
valid_pct=0.2, bs=2**8,
N_save=N, T_save=400,
)model_args = dict(# VAE ###########################
o_dim=ds_args['T']-1,
nc_in=1, nc_out=6,
nf=[16]*4,
avg_size=16,
encoder=[200,100],
z_dim=6,
decoder=[100,200],
beta=0,
# WaveNet ########
in_channels=1,
res_channels=16,skip_channels=16,
c_channels=6,
g_channels=0,
res_kernel_size=3,
layer_size=4, # 6 # Largest dilation is 2**layer_size
stack_size=1,
out_distribution= "Normal",
num_mixtures=1,
use_pad=False,
model_name = 'SPIVAE',
)dls = load_data(ds_args)model = VAEWaveNet(**model_args).to(DEVICE)loss_fn = Loss(model.receptive_field, model.c_channels,
beta=model_args['beta'], reduction='mean')learn = Learner(dls, model, loss_func=loss_fn,)E=4learn.fit_one_cycle(E, lr_max=1e-4)| epoch | train_loss | valid_loss | time |
|---|---|---|---|
| 0 | 0.982180 | 0.949590 | 00:28 |
| 1 | 0.934559 | 0.880010 | 00:25 |
| 2 | 0.882010 | 0.822553 | 00:26 |
| 3 | 0.843744 | 0.810182 | 00:29 |